Command Palette
Search for a command to run...
MediCLIP: Adapting CLIP for Few-shot Medical Image Anomaly Detection
MediCLIP: Adapting CLIP for Few-shot Medical Image Anomaly Detection
Ximiao Zhang Min Xu Dehui Qiu Ruixin Yan Ning Lang Xiuzhuang Zhou
Abstract
In the field of medical decision-making, precise anomaly detection in medical imaging plays a pivotal role in aiding clinicians. However, previous work is reliant on large-scale datasets for training anomaly detection models, which increases the development cost. This paper first focuses on the task of medical image anomaly detection in the few-shot setting, which is critically significant for the medical field where data collection and annotation are both very expensive. We propose an innovative approach, MediCLIP, which adapts the CLIP model to few-shot medical image anomaly detection through self-supervised fine-tuning. Although CLIP, as a vision-language model, demonstrates outstanding zero-/fewshot performance on various downstream tasks, it still falls short in the anomaly detection of medical images. To address this, we design a series of medical image anomaly synthesis tasks to simulate common disease patterns in medical imaging, transferring the powerful generalization capabilities of CLIP to the task of medical image anomaly detection. When only few-shot normal medical images are provided, MediCLIP achieves state-of-the-art performance in anomaly detection and location compared to other methods. Extensive experiments on three distinct medical anomaly detection tasks have demonstrated the superiority of our approach. The code is available at https://github.com/cnulab/MediCLIP.
One-sentence Summary
The authors from Capital Normal University, Peking University Third Hospital, and Beijing University of Posts and Telecommunications propose MediCLIP, a novel few-shot medical image anomaly detection framework that adapts CLIP via self-supervised fine-tuning with multi-task synthetic anomaly generation, enabling state-of-the-art performance with minimal normal training data and achieving strong zero-shot generalization across diverse medical imaging tasks.
Key Contributions
- Few-shot medical image anomaly detection is a critical yet underexplored task due to the high cost and scarcity of annotated medical data; MediCLIP addresses this by enabling accurate anomaly detection and localization using only a few normal images per task, without requiring any anomaly images or pixel-level labels.
- MediCLIP adapts the CLIP model through self-supervised fine-tuning with a novel set of medical image anomaly synthesis tasks that simulate diverse disease patterns, while leveraging learnable prompts and adapters to enable effective multi-scale lesion localization and robust generalization.
- Extensive experiments on three medical datasets—CheXpert, BrainMRI, and BUSI—show that MediCLIP achieves state-of-the-art performance, outperforming existing methods by approximately 10% and reaching 94% of the full-shot SQUID model’s accuracy on CheXpert while using less than 1% of the training data.
Introduction
In medical image analysis, accurate anomaly detection is critical for clinical decision-making, yet most existing methods rely on large annotated datasets, making development costly and impractical for rare conditions. Prior approaches often require extensive normal data and struggle with few-shot scenarios where only a handful of normal images are available, limiting real-world deployment. The authors introduce MediCLIP, a novel framework that adapts the CLIP vision-language model for few-shot medical anomaly detection by leveraging self-supervised fine-tuning with synthetic anomaly images. To overcome the lack of real anomaly data, they design medical-specific anomaly synthesis tasks that simulate diverse disease patterns, enabling the model to learn robust anomaly representations. MediCLIP further enhances localization through learnable prompts and adapter modules that align vision and text features across multiple scales. Evaluated on three medical imaging datasets—CheXpert, BrainMRI, and BUSI—MediCLIP achieves state-of-the-art performance with up to 10% improvement over existing methods, matching 94% of full-shot model performance while using less than 1% of the training data. Notably, it demonstrates strong zero-shot generalization across tasks, suggesting potential as a unified diagnostic tool.
Dataset
- The dataset comprises three medical imaging sources: Stanford CheXpert (chest X-rays), BrainMRI (2D brain MRIs), and BUSI (breast ultrasound images).
- CheXpert includes 12 disease categories with clinical chest X-rays; BrainMRI contains both normal and tumor-affected brain scans; BUSI features breast ultrasound images labeled as normal, benign, or malignant, with pixel-level disease annotations.
- All disease cases are treated as anomalies. For training, the authors use k = {4, 8, 16, 32} normal images per dataset.
- Test sets consist of 250 normal and 250 anomaly images for CheXpert, 65 normal and 155 anomaly images for BrainMRI, and 101 normal and 647 anomaly images for BUSI.
- The model uses a ViT-L/14 CLIP backbone, extracting features from the 12th, 18th, and 24th layers of the visual encoder.
- A learnable token count of M = 8 and a linear adapter are used, with a temperature parameter τ set to 0.07.
- Anomaly synthesis employs three tasks—CutPaste, GaussIntensityChange, and Source—each applied with equal probability.
- For CutPaste and GaussIntensityChange, Perlin noise generates anomalous shapes, which are binarized into masks. The Source task uses randomly sized and rotated ellipses or rectangles as masks.
- Intensity variation (γ) is sampled uniformly from [-0.6, -0.4) ∪ [0.4, 0.6), and the blending factor (α) from [√2, 4).
- All input images are resized to 224 × 224 pixels.
- Experiments are repeated three times with different randomly sampled support sets to ensure robustness.
Method
The authors leverage a modified CLIP architecture, referred to as MediCLIP, to address few-shot anomaly detection in medical imaging. The overall framework, illustrated in the figure below, integrates learnable prompts, adapters, and a multi-task anomaly synthesis strategy to adapt the vision-language model for detecting and localizing anomalies with minimal labeled data.
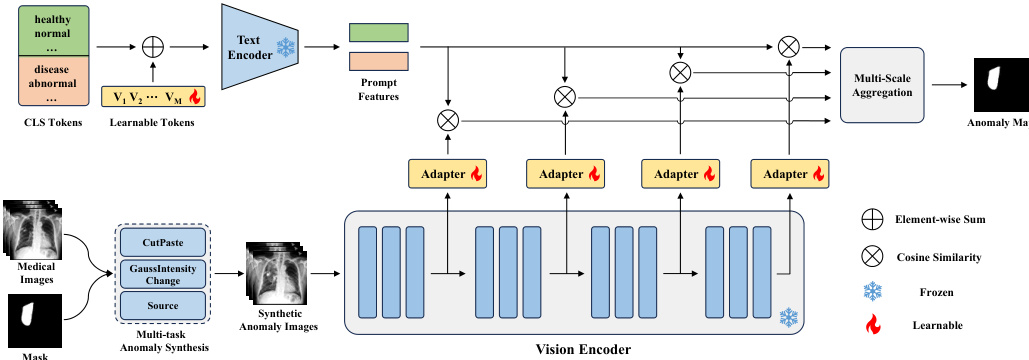
The framework begins with the construction of learnable prompts to represent normal and anomalous classes. Instead of relying on manually designed prompts, the authors employ learnable word embeddings for the prompt tokens, denoted as [V1],[V2],...,[VM], followed by a fixed class token [CLS]. For normal cases, class tokens such as [healthy] and [normal] are used, while [disease] is used for anomalies. The text encoder of CLIP, F(⋅), processes these prompts to generate feature representations, F(p), which are then averaged to obtain mean feature representations fn and fa for normal and anomalous classes, respectively.
To adapt the vanilla CLIP model for anomaly detection, the authors introduce a set of learnable adapters, ϕj(⋅), which are inserted into the vision encoder at multiple intermediate layers. The vision encoder, Gj(⋅), extracts multi-scale visual features, Gj(X^), from a synthetic anomaly image X^. Each adapter projects the feature map Gj(X^) to a feature space of the same dimension as the prompt features, resulting in gj∈RHj×Wj×C. The similarity between the projected visual features gj(h,w) and the mean prompt features fn and fa is computed using cosine similarity, normalized by a temperature parameter τ, to generate per-location similarity scores Snj(h,w) and Saj(h,w). This process yields multi-scale similarity matrices for both normal and anomalous classes.
The framework aggregates these multi-scale similarity matrices by upscaling them to the original image resolution H×W and computing their average to produce final similarity maps Sn and Sa. The loss function for training combines Focal loss and Dice loss, applied to the concatenated similarity maps and the anomaly mask Y, to optimize the learnable parameters in the prompts and adapters. During inference, the anomaly synthesis module is removed, and the query image is directly processed by the vision encoder. The anomaly map is derived from Sa, and the image-level anomaly score is determined by the maximum value in Sa.
The synthetic anomaly images are generated through a multi-task anomaly synthesis strategy, which includes CutPaste, GaussIntensityChange, and Source. CutPaste simulates misplacement anomalies by pasting a randomly selected image patch to a new location using Poisson image editing. GaussIntensityChange models density variations by altering pixel intensities within a mask using Gaussian noise. The Source task simulates deformations by repelling pixels within the mask from a central point, effectively modeling proliferative anomalies. These synthesis tasks are designed to generate diverse and realistic anomaly patterns, enhancing the model's ability to generalize to unseen anomalies.
Experiment
- MediCLIP outperforms other methods by approximately 10% in few-shot anomaly detection across three datasets, demonstrating superior generalization through CLIP-based anomaly synthesis.
- On the BUSI dataset, MediCLIP achieves more accurate anomaly localization than baseline methods, with visualizations confirming precise identification of lesion regions.
- MediCLIP achieves strong zero-shot performance across diverse datasets, validating its generalization capability via a multi-task anomaly synthesis strategy.
- Ablation studies confirm the necessity of the learnable token and adapter, with their removal leading to performance degradation.
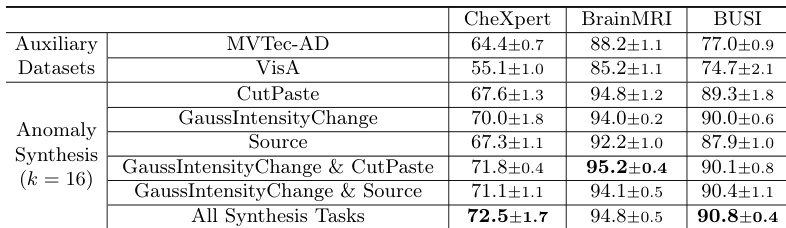
- Training with auxiliary industrial datasets (MVTec-AD, VisA) improves performance, but medical anomaly synthesis yields better generalization, and combining multiple tasks further enhances results.
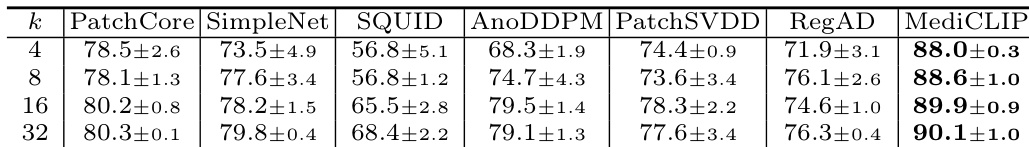
The authors use an ablation study to evaluate the impact of learnable token and adapter components in MediCLIP, showing that their removal significantly reduces performance. Results indicate that MediCLIP achieves the highest Image-AUROC scores across all support set sizes, with performance improving as the support set size increases, reaching 90.1% at k=32.
The authors use Table 4 to evaluate the impact of the learnable token and adapter in MediCLIP, showing that both components are essential for optimal performance. Results indicate that replacing the learnable token with hard prompt templates or using average pooling instead of the adapter significantly reduces Image-AUROC, demonstrating their irreplaceable role in the model.
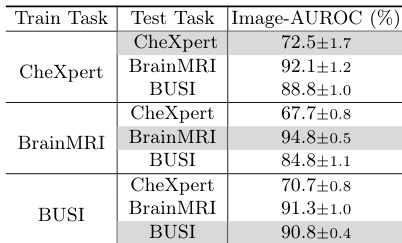
Results show that the combination of learnable token and adapter in MediCLIP significantly improves anomaly detection performance across all datasets, with the best results achieved when both components are used. The ablation study demonstrates that neither the learnable token nor the adapter can be replaced without a substantial drop in performance, highlighting their essential roles in the model's effectiveness.
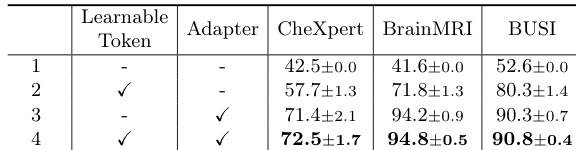
The authors use an ablation study to evaluate the impact of learnable token and adapter components in MediCLIP, showing that their removal significantly reduces performance across all datasets. Results indicate that the learnable token and adapter are essential for achieving high Image-AUROC scores, with MediCLIP outperforming all other configurations in the study.
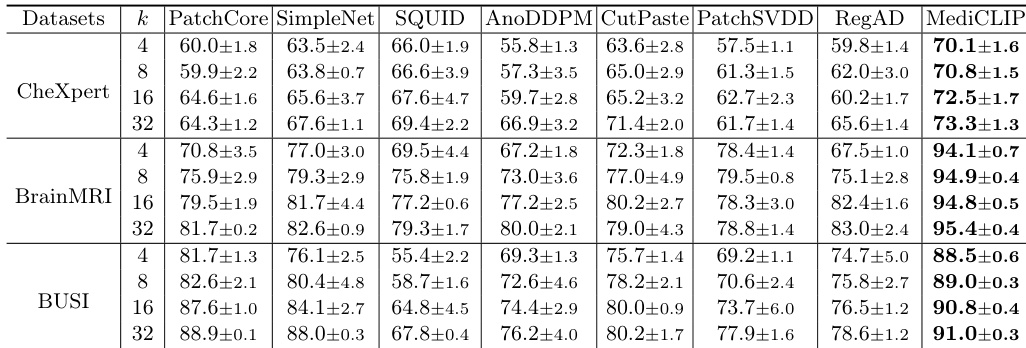
The authors use an ablation study to evaluate the impact of different anomaly synthesis tasks in MediCLIP, showing that combining multiple tasks significantly improves performance across all datasets. Results show that the full set of synthesis tasks achieves the highest Image-AUROC scores, with 72.5±1.7 on CheXpert, 94.8±0.5 on BrainMRI, and 90.8±0.4 on BUSI, outperforming individual or fewer task combinations.